OpenMS In KNIME
As part of the Center for Integrative Bioinformatics (CiBi) in the German Network for Bioinformatics (deNBI), we are currently focusing our development efforts on the integration of OpenMS into KNIME. KNIME is a well-established data analysis framework that supports the generation of workflows for data analysis. Using a Common Tool Description (CTD) file which is writeable by every TOPP tool and a node generator program (Generic Knime Nodes), all TOPP tools can be made available to run in KNIME.
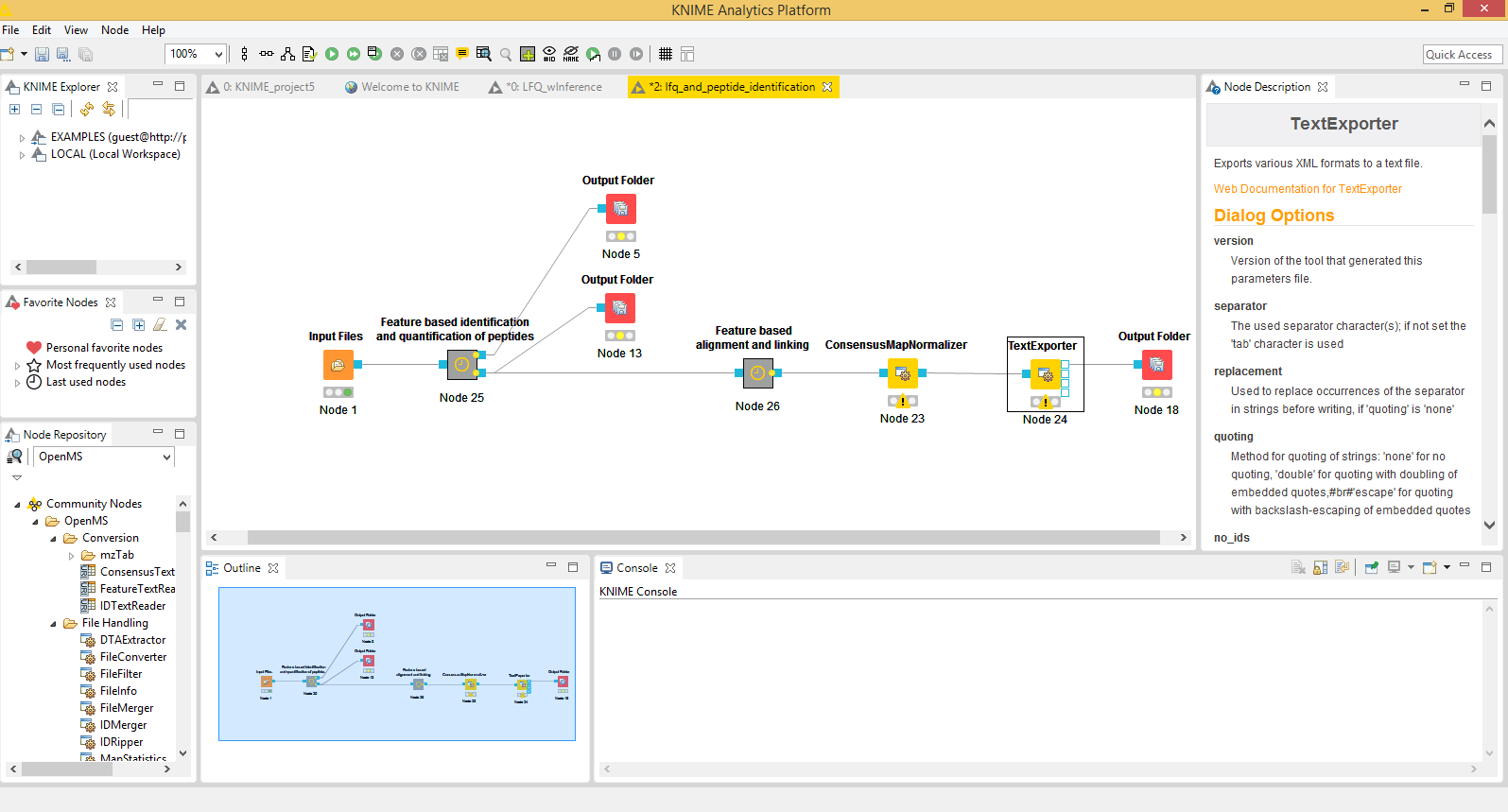
Example workflow in KNIME
Installing OpenMS in KNIME
Installation of OpenMS in KNIME is very easy and platform-independent across Windows, MacOSX and Linux. Download the latest KNIME release from the KNIME website. If you choose the full install of KNIME you most likely can skip the following installation routine since all required plugins should be installed by default. If you chose the standard (core) installation, follow the instructions here or in the extended User Tutorial :
- In KNIME click on Help
- Install new Software.
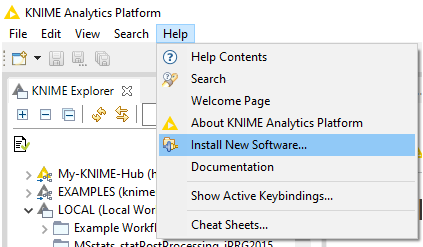
Example workflow in KNIME
- Install the required KNIME File Handling nodes from the official KNIME Update Site (a standard entry in the update sites). Choose the update site from the “Work with:” dropdown menu.
Name: KNIME Analytics Platform ${YOUR_KNIME_VERSION} Update Site Location: http://update.knime.org/analytics-platform/${YOUR_KNIME_VERSION}
Filter the results for “File handling” and select the KNIME File Handling Nodes. Click Next and install.
Next, we will install the actual OpenMS plugin. Next to the “Work with:” dropdown menu, click on “Add…”.
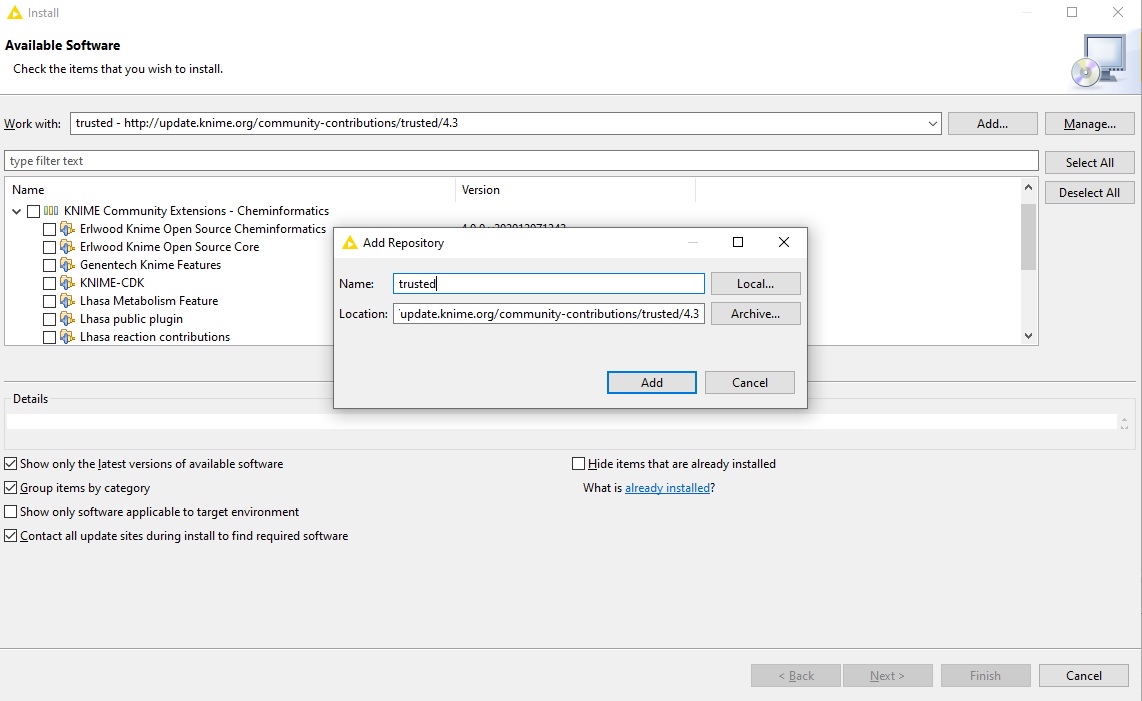
In the opening dialog fill in at least one of the following additional Update Sites (if not already present):
Recommended:
Name: KNIME Community Contributions (Stable)
Location: http://update.knime.org/community-contributions/trusted/${YOUR_KNIME_VERSION}
Unstable:
Name: KNIME Nightly Community Contributions (trunk)
Location: http://update.knime.org/community-contributions/trunk
Use the search or navigate to “KNIME Community Contributions – Bioinformatics & NGS” and select “OpenMS”. Then click “Next” and follow the installation instructions. A restart of KNIME might be necessary afterward. On Windows, you might be prompted to install additional requirements like the Microsoft Visual Studio Redistributable for the conversion software ProteoWizard that is packaged with our plugin. After a restart of KNIME the OpenMS nodes will be available in your Node Repository (panel on the lower left) under “Community Nodes”.
Creating workflows with KNIME
Download our Introduction to OpenMS in KNIME containing hands-on training material covering also basic usage of KNIME. See the official KNIME Getting Started guide for a more in-depth view of the KNIME functionality besides OpenMS.
When you encounter any problems, please see our Support page and specifically for the usage of OpenMS in KNIME, the KNIME community contributions forum. When in doubt where to turn to, open an issue on our GitHub issue tracker.
Creating your own Generic KNIME Nodes
See the more advanced instructions on our Getting Started page for developers.
Example
Please click on the link here to check some emxaple workflows.
OpenMS In GALAXY
Nodes for OpenMS tools have been kindly made available via the PROTEOMICS, METABOLOMICS, CHEMISTRY section at https://usegalaxy.eu/ and can be browsed by searching for OpenMS in the tools search box.

OpenMS In Nextflow
SCALABLE NF-CORE COMPATIBLE NEXTFLOW PIPELINES
Click on “Launch” to configure the pipeline for your data online and launch it via nextflow’s tower app (by registering a compute environment there) or by copying a configuration token for your local computer or HPC head node.