Note
Go to the end to download the full example code. or to run this example in your browser via Binder
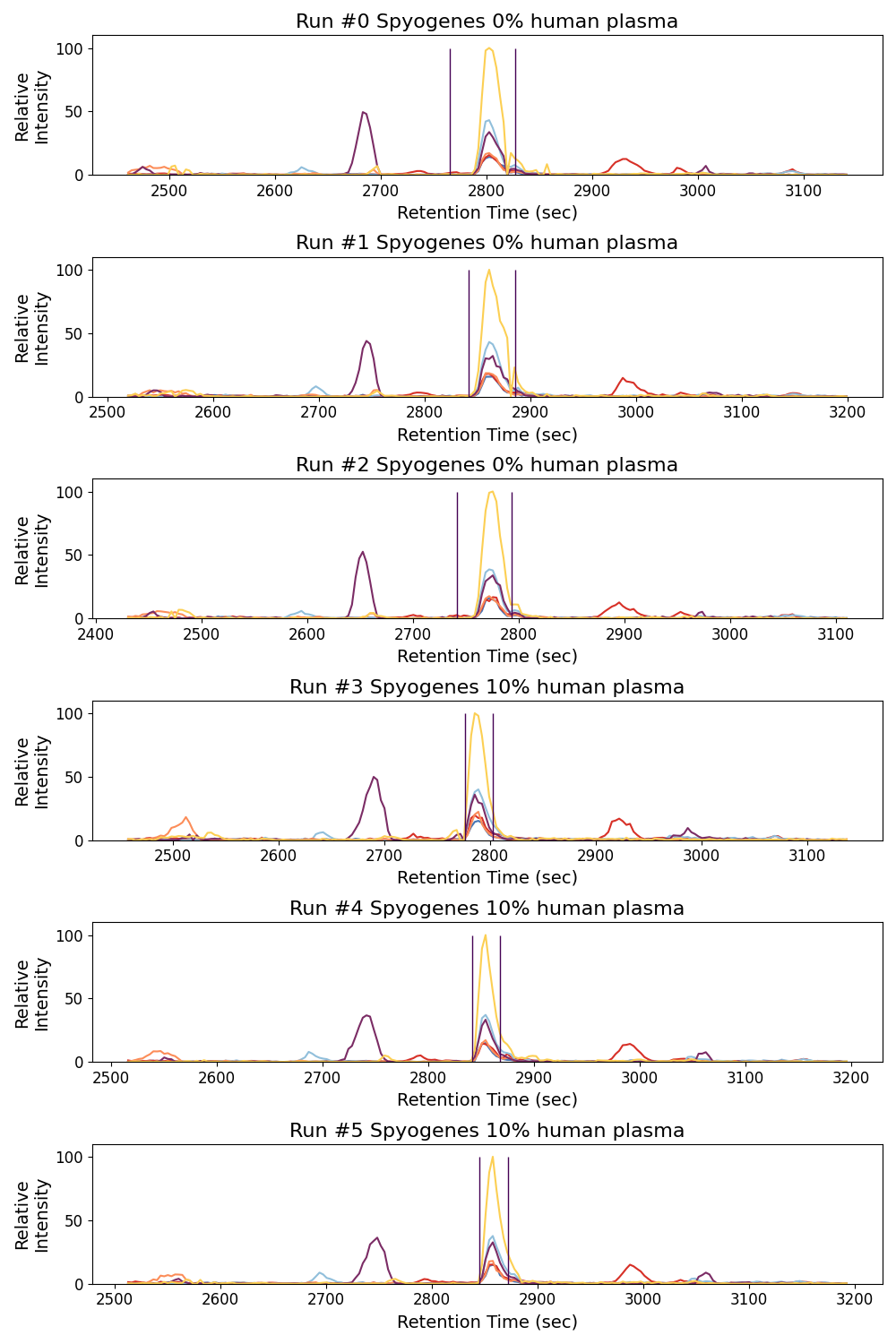
Plot Spyogenes subplots ms_matplotlib#
Here we show how we can plot multiple chromatograms across runs together
Downloading spyogenes.zip...
Downloaded spyogenes.zip successfully.
Unzipped files successfully.
<Figure size 1000x1500 with 6 Axes>
import pandas as pd
import requests
import zipfile
import numpy as np
import matplotlib.pyplot as plt
pd.options.plotting.backend = "ms_matplotlib"
###### Load Data #######
# URL of the zip file
url = "https://github.com/OpenMS/pyopenms_viz/releases/download/v0.1.3/spyogenes.zip"
zip_filename = "spyogenes.zip"
# Download the zip file
try:
print(f"Downloading {zip_filename}...")
response = requests.get(url)
response.raise_for_status() # Check for any HTTP errors
# Save the zip file to the current directory
with open(zip_filename, "wb") as out:
out.write(response.content)
print(f"Downloaded {zip_filename} successfully.")
except requests.RequestException as e:
print(f"Error downloading zip file: {e}")
except IOError as e:
print(f"Error writing zip file: {e}")
# Unzipping the file
try:
with zipfile.ZipFile(zip_filename, "r") as zip_ref:
# Extract all files to the current directory
zip_ref.extractall()
print("Unzipped files successfully.")
except zipfile.BadZipFile as e:
print(f"Error unzipping file: {e}")
annotation_bounds = pd.read_csv(
"spyogenes/AADGQTVSGGSILYR3_manual_annotations.tsv", sep="\t"
) # contain annotations across all runs
chrom_df = pd.read_csv(
"spyogenes/chroms_AADGQTVSGGSILYR3.tsv", sep="\t"
) # contains chromatogram for precursor across all runs
##### Set Plotting Variables #####
pd.options.plotting.backend = "ms_matplotlib"
RUN_NAMES = [
"Run #0 Spyogenes 0% human plasma",
"Run #1 Spyogenes 0% human plasma",
"Run #2 Spyogenes 0% human plasma",
"Run #3 Spyogenes 10% human plasma",
"Run #4 Spyogenes 10% human plasma",
"Run #5 Spyogenes 10% human plasma",
]
fig, axs = plt.subplots(len(np.unique(chrom_df["run"])), 1, figsize=(10, 15))
# plt.close ### required for running in jupyter notebook setting
# For each run fill in the axs object with the corresponding chromatogram
plot_list = []
for i, run in enumerate(RUN_NAMES):
run_df = chrom_df[chrom_df["run_name"] == run]
current_bounds = annotation_bounds[annotation_bounds["run"] == run]
run_df.plot(
kind="chromatogram",
x="rt",
y="int",
grid=False,
by="ion_annotation",
title=run_df.iloc[0]["run_name"],
title_font_size=16,
xaxis_label_font_size=14,
yaxis_label_font_size=14,
xaxis_tick_font_size=12,
yaxis_tick_font_size=12,
canvas=axs[i],
relative_intensity=True,
annotation_data=current_bounds,
xlabel="Retention Time (sec)",
ylabel="Relative\nIntensity",
annotation_legend_config=dict(show=False),
legend_config={"show": False},
)
fig.tight_layout()
fig
Total running time of the script: (0 minutes 1.158 seconds)
